Nontargeted Metabolite Profiling in Next-Generation Plant Breeding: A Case Study in Malting Barley
Special Issues
The power of nontargeted metabolite profiling is illustrated in a study focused on the determination of molecular markers in malting barley that are predictive of desirable malting quality for brewing applications. The metabolite extraction, detection, and analysis methods are highthroughput and reproducible, and therefore, this approach represents a practical addition to the plant breeder’s molecular toolbox.
Nontargeted metabolite profiling by ultrahigh-pressure liquid chromatography–mass spectrometry (UHPLC-MS) is a powerful technique to investigate the influence of genetic and environmental factors on metabolic phenotypes in plants. The approach offers an unbiased and in-depth analysis that can reveal molecular markers of desirable phenotypic traits that can be complementary to genetic markers in plant breeding efforts. Here, the power of nontargeted metabolite profiling is illustrated in a study focused on the determination of molecular markers in malting barley that are predictive of desirable malting quality for brewing applications.
Plant breeding has been central to agriculture for thousands of years, and plant research continues to be a critical component of global agriculture for food production. Each year, breeders release new plant varieties with improved traits. Researchers continually seek to improve traits such as crop yield and pathogen and pest resistance as well as other qualities that are important to downstream food storage and processing.
The major goal in plant breeding is to develop a new variety (also known as a line or cultivar) with a set of genes that result in an optimal suite of phenotypes in a particular environment. Plant breeding is largely centered on improving crop yield (for example, kilograms of plant tissue per hectare), but it is also focused on traits that improve an amenability to harvest (such as uniform plant height), adaptation to local environments (such as temperatures, photoperiod), and growth habit (such as plant morphology traits that lead to a specific canopy structure and higher density). In addition to agronomic traits, there is a significant effort to breed plants for downstream quality in food or industrial traits. At its core, crop quality is synonymous with a specific chemical composition of the plant. Through traditional techniques and years of work, breeders can fine-tune the chemical composition of their crop to develop specific colors, flavors, extended storage, and compatibility with food processing techniques such as cooking or baking.
New plant varieties are developed by crossing two plants to produce offspring (sometimes hundreds of offspring per cross), each of which contains a unique mixture of genes from the two parents. This genetic shuffling results in an array of phenotypes and, after many cycles of further crossing (by self or sibling pollination to reduce heterogeneity), a population of plants is evaluated to identify which plant lines have superior performance over the initial parents. The entire process, from the initial cross to final release, can take longer than 10 years for some crops and relies on extensive field trials performed at many locations.
Given the large size of a plant breeding population, the breadth of phenotypes of interest, and the many years required to release new varieties to farmers, next-generation plant breeding seeks to incorporate high-throughput molecular techniques to speed up the process while reducing costs. In this regard, the molecular “omics” technologies have been promising, specifically for the development of genomic selection models and the identification of genetic loci that regulate gene expression (1). The three most widely used omics-genomics, transcriptomics, and proteomics-use detection and analysis techniques that are fine-tuned for a specific chemical structure, specifically DNA, RNA, and proteins. In contrast, metabolomics, a fourth omics approach defined by the detection and analysis of small molecules in a biological sample, requires analytical techniques that can quantify a highly diverse suite of small molecules. Mass spectrometry (MS) coupled to either liquid chromatography (LC) or gas chromatography (GC) separation has been a major technological driver in metabolomics research for its ability to detect a broad range of compounds at varying abundances.
In plants, the significant breadth of compounds that can exist among different tissues (leaf, stem, root) and among species creates unique challenges in a metabolomics experiment. For example, a single systems-level experiment in plant metabolism can require multiple independent analyses to effectively profile organic acids, phospholipids, amines, amino acids, terpenes, sterols, saccharides, and oxylipins. Furthermore, plants produce secondary or specialized metabolites, many of which function to combat external abiotic or biotic stresses (2). The Dictionary of Natural Products website has references for more than 250,000 compounds (3), and approximately 25,000 of those are terpenes, often with specialized function (4). To add to this complexity, plants often further modify secondary compounds in response to external stresses. For example, glycosylation allows for the transport of a molecule to a specific plant cell organelle and methylation allows for metabolites to volatilize and generate signals to neighboring plant tissues or other plants.
Here, we describe a study published in the Plant Biotechnology Journal that investigated the utility of LC–MS metabolomics as a next-generation analytical tool in plant breeding (5). The experiment characterized the relationship between target traits in malting barley and the barley and malt metabolome. Malting barley, a food crop developed for the brewing industry, is an ideal plant system because of the close relationship between the chemical composition of the grain and the final product: beer. Like other food crops, malting barley has been bred for high yield and is well-adapted to many growing environments. In addition, malting quality is defined by chemical traits in the barley grain that are critical to the brewing industry (Table I). Two example traits include malt extract and free amino nitrogen, which are quantitative values that represent the amount of carbon (saccharides) and nitrogen (amino acids and small peptides) available for microbial fermentation. Other barley grain traits important to brewing include oligosaccharides (plant cell wall components) and enzyme content to break down oligosaccharides.
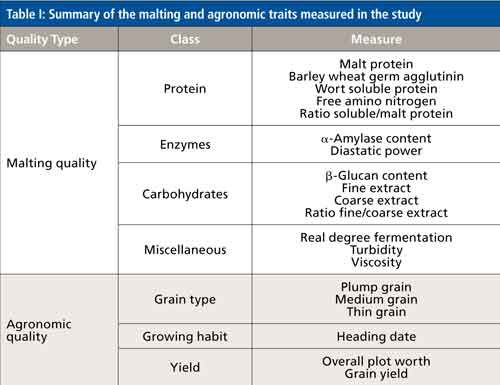
The study used a nontargeted metabolomics analytical workflow coupled with a novel chemo-informatics approach to interpret the spectral data and annotate metabolites. Multivariate statistics were also applied to explain the relationships between barley and malt metabolites with malting quality traits. These relationships can be the foundation to apply small-molecule profiling to the plant breeding toolbox.
Experimental Design
The overall goal of the experiment was to determine if variation in plant traits evaluated by a plant breeder (Vg, genetic variation and Ve, environmental variation) are associated with metabolite variation in the barley grain (Figure 1). Barley is classified as “two-row” or “six-row” based on the number of grain rows that occur in the barley head. The two barley types have been kept in distinct breeding pools and are nearly subspecies. This breeding history has led to two-row types having a higher content of malt extract (that is, soluble, fermentable small molecules such as saccharides) and six-row types having more protein (historically used as animal feed). Specifically, we aimed to determine if metabolite variation in the barley grain is consistent with the genetic (two-row versus six-row) and environmental variation that would normally be observed by the breeder. In our study, the two-row and six-row types were grown in two locations: Idaho and Montana. The breeder evaluated 20 traits (both agronomic and malting quality) and principal component analysis (PCA) revealed both row-type and environment contributed to variation in overall quality (Figure 2a).
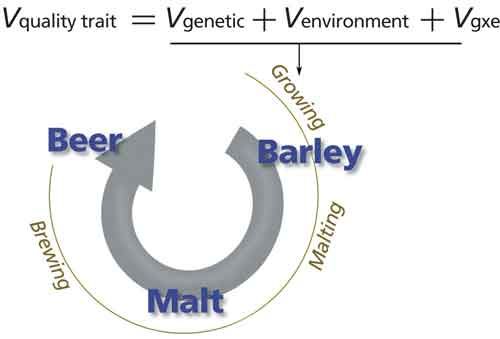
Figure 1: Schematic illustrating the contribution of genetic and environmental variation on barley and malt.
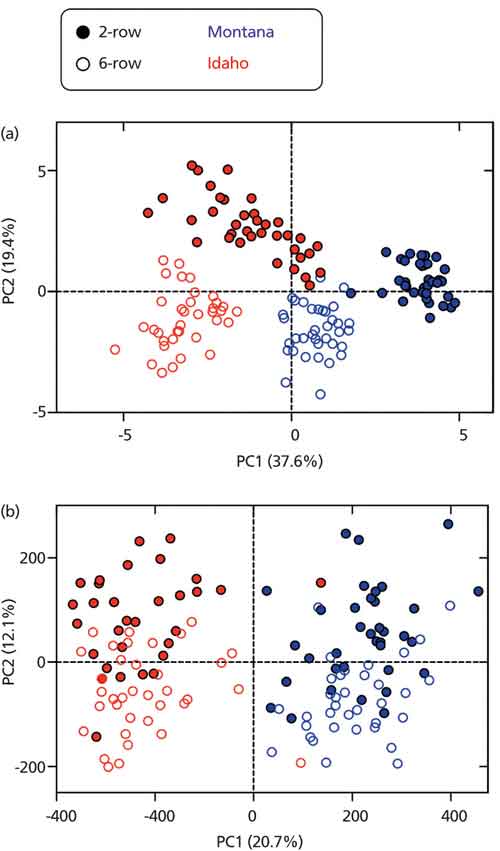
Figure 2: Principal component analysis scores plot for (a) barley and (b) malt metabolites. Adapted with permission from reference 5.
Metabolite detection for barley and malt samples was performed using ultrahigh-pressure liquid chromatography (UHPLC) coupled to time-of-flight mass spectrometry (TOF-MS) and peak detection and quantitation was performed using open source software (XCMS) (6). A nontargeted workflow for plant metabolomics was used (Figure 3). While previous studies have described the use of targeted metabolomics (that is, focused on a predetermined panel of molecules), this study represents one of the first nontargeted (that is, unbiased) investigation of metabolism in the context of plant breeding.
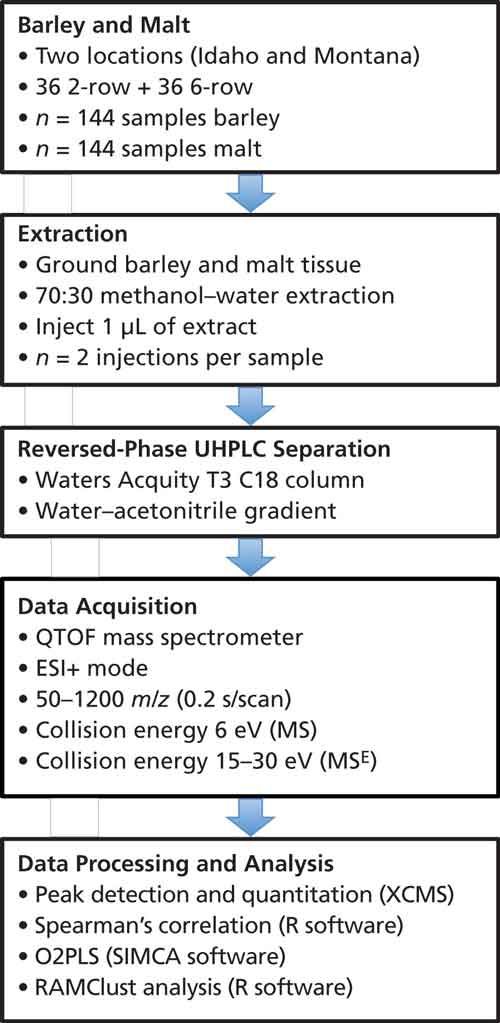
Figure 3: Schematic illustrating the experimental workflow used to detect and quantify metabolites in barley and malt.
The resulting malting barley metabolite data set consisted of 27,420 molecular features. The metabolomics data was evaluated using PCA, which characterized the profiles for both barley and malt (Figure 2). The results showed that the barley metabolome was mostly influenced by environment, whereas the malt metabolome was highly influenced by both genotype and environment. Thus, while brewers have long known the importance of carefully selecting a two- or six-row malt, the metabolite differences between the two barley types are in fact exacerbated by the malting process.
The data was then investigated to determine if there are direct links to the plant phenotypic traits evaluated by the breeder. An O2PLS model was used, which is a variation of orthoganol projection to latent structures (OPLS) (7). In this model, the 20 phenotypic traits were regressed against the quantitative metabolite data (y = 20 plant phenotypes, x = 27,420 metabolites). The output was a model with four components; in other words, four distinct relationships between metabolites and quality traits (Figure 4). The O2PLS loadings, which represent the contribution of a specific metabolite to the four components, was used to seek out compounds that were important to breeding. Finally, a z-score was calculated for each metabolite relative to the four components and resulted in a set of 216 metabolites that were significantly associated with at least one of the 20 quality traits.
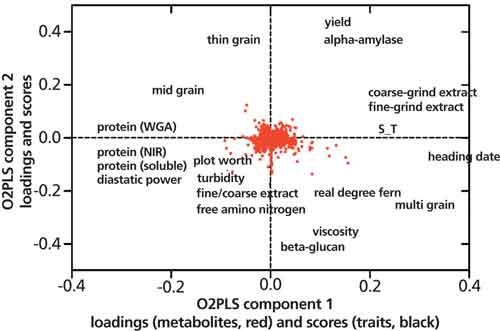
Figure 4: O2PLS overview plot of showing associations of metabolites (red dots) to traits.
Chemo-Informatics to Annotate Important Plant Compounds for Breeding
The annotation of mass spectrometry data as biologically relevant metabolites remains a bottleneck in metabolomics. One of the reasons this is so difficult is that each metabolite is actually represented by multiple peaks in the data. For example, molecules can lose water or form adducts and dimers. These events create a complexity that is difficult to deconvolute in complex samples. Recently, our group developed a novel method, called RAMClust, for grouping mass spectral features by exploiting the inherent variation in a metabolomics experiment (8). That is, if the quantitative values of two spectral peaks are correlated across every injection made in the experiment and the peaks coelute in chromatographic time, then it is likely that they are derived from the same molecule. The RAMClust algorithm was applied to the barley and malt metabolomics dataset to generate peak groups (spectra) for the 216 metabolites that were determined to be important for breeding. These spectra were screened against our in-house spectral library and external metabolites databases and we were able to annotate many amines, amino acids, purines, and lipids. The power of the RAMClust algorithm is illustrated in the ability to distinguish two metabolites that completely coeluted but had different quantitative variation across the dataset. In this case, we were able to annotate two very different barley compounds, a tannin and a barley hordatine (an alkaloid) (Figure 5) that would have otherwise remained as unknowns.
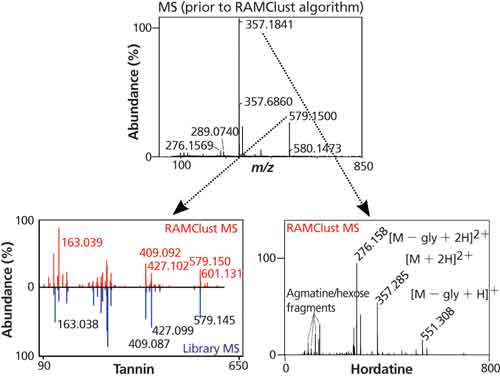
Figure 5: Application of the RAMClust algorithm enabled differentiation of coeluted m/z values as two distinct metabolites. Top panel: unprocessed mass spectrum. Bottom panels: RAMClust generated spectra for m/z values 579.150 (left - compared to spectral library entry) and 357.184 (right). Adapted with permission from reference 5.
Conclusion
Taken together, the results from this study demonstrate the utility of nontargeted metabolite profiling for the identification of metabolites that are associated with the agronomic and quality traits normally evaluated by a breeder. The 216 metabolites identified in this study can serve as markers for agronomic and quality traits in malting barley. It is important to note that the metabolite extraction, detection, and analysis methods are high-throughput and reproducible, and therefore, this approach represents a valuable and practical addition to the plant breeder’s molecular toolbox. Using chromatography coupled with mass spectrometry to evaluate metabolites in a breeding population can facilitate the rapid evaluation of barley breeding lines and reduce the time required for variety development.
Acknowledgment
This study was funded by the American Malting Barley Association. We would like to thank Gary Hanning and his team at AB-Inbev for materials and guidance that were critical to this study.
This article was originally published in the December 2015 issue of LCGC North America as an installment of the “MS–The Practical Art” column and was edited by LCGC columnist Kate Yu (9).
References
- J.-L. Jannink, A.J. Lorenz, and H. Iwata, Briefings Funct. Genomics9, 166–177 (2010).
- A.L. Schilmiller, E. Pichersky, and R.L. Last, Curr. Opin. Plant Biol.15, 338–344 (2012).
- Available at: dnp.chemnetbase.com.
- J. Gershenzon and N. Dudareva, Nat. Chem. Biol.3, 408–414 (2007).
- A.L. Heuberger, C.D. Broeckling, K.R. Kirkpatrick, and J.E. Prenni, Plant Biotechnol. J.12, 147–160 (2014).
- C.A. Smith, E.J. Want, G. O’Maille, R. Abagyan, and G. Siuzdak, Anal. Chem.78(3), 779–787 (2006).
- M. Bylesjo, D. Eriksson, M. Kusano, T. Moritz, and J. Trygg, Plant Journal52, 1181–1191 (2007).
- C.D. Broeckling, F.A. Afsar, S. Neumann, A. Ben-Hur, and J.E. Prenni, Anal. Chem.86(14), 6812–6817 (2014).
- A. Heuberger, C. Broeckling, and J. Prenni, LCGC North Am.33(12), 910–913, 917 (2015).
Adam Heuberger is an Assistant Professor at Colorado State University with more than 8 years in plant, animal, and microbial metabolomics. His research applies nontargeted metabolite profiling to improve quantity and quality traits in food crops.
Corey D. Broeckling is the Associate Director of the Proteomics and Metabolomics Facility and an Assistant Professor at Colorado State University. He has more than 13 years of experience in metabolomics and chemoinformatics.
Jessica Prenni has more than 13 years of experience with biological mass spectrometry and is the Director of the Proteomics and Metabolomics Facility and Associate Professor at Colorado State University. Direct correspondence to SpectroscopyEdit@ubm.com

Synthesizing Synthetic Oligonucleotides: An Interview with the CEO of Oligo Factory
February 6th 2024LCGC and Spectroscopy Editor Patrick Lavery spoke with Oligo Factory CEO Chris Boggess about the company’s recently attained compliance with Good Manufacturing Practice (GMP) International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use (ICH) Expert Working Group (Q7) guidance and its distinction from Research Use Only (RUO) and International Organization for Standardization (ISO) 13485 designations.