Using Thermostable Raman Interaction Profiling (TRIP) For Protein Binding Screening
Proteins are vital components in the functioning of living organisms, and what they do depends on various interactions they have with other molecules. A new technique, called thermostable Raman interaction profiling (TRIP), can be used to analyze the sensitive detection of binding between proteins and ligands at low concentrations and low doses under conditions that mimic the body's natural environment (1). TRIP has significant implications for drug development, providing a valuable tool for researchers and pharmaceutical companies. It opens possibilities for high-throughput drug screening and real-time monitoring of the interactions between proteins and drugs (1).
Narangerel Altangerel (pictured), Zhenhuan Yi, and Marlan Scully of Texas A&M University recently used TRIP to analyze eight protein–ligand systems for a study published in PNAS. Spectroscopy recently spoke to these three researchers about their findings and what the implications are for high-throughput drug screening. The research team collaborated on the responses, which have been lightly edited for style and clarity.
Dr. Narangerel Altangerel of Texas A&M University | Image Credit: © Narangerel Altangerel

Can you provide a brief overview about why studying protein interactions is important, especially it comes to drug development applications?
Proteins are the workhorses of biological systems, responsible for carrying out various functions within cells. Many diseases, such as cancer, neurodegenerative disorders, and infectious diseases, are often the result of dysregulated protein interactions. Understanding these interactions can provide crucial insights into the underlying mechanisms of disease. Identifying and characterizing protein interactions can lead to the discovery of potential drug targets. By targeting specific proteins or disrupting harmful interactions, researchers can develop drugs to treat diseases. For example, many cancer drugs target proteins involved in aberrant cell signaling pathways.
A simple, label-free screening technique capable of directly sensing interaction-in-solution involving large proteins, such as antibodies, holds promise for an important tool in the field of drug development.
Your study presents a thermostable Raman interaction profiling (TRIP) technique to screen protein binding (1). Why was this technique best for the analysis that you were conducting?
Thermostable Raman Interaction Profiling (TRIP) represents a promising innovation in the field of molecular interaction studies and drug discovery. Its ability to directly measure interactions in physiologically relevant conditions, its label-free nature, and its compatibility with aqueous samples make it a valuable tool for researchers seeking to better understand and develop treatments for various diseases by targeting specific protein-ligand interactions.
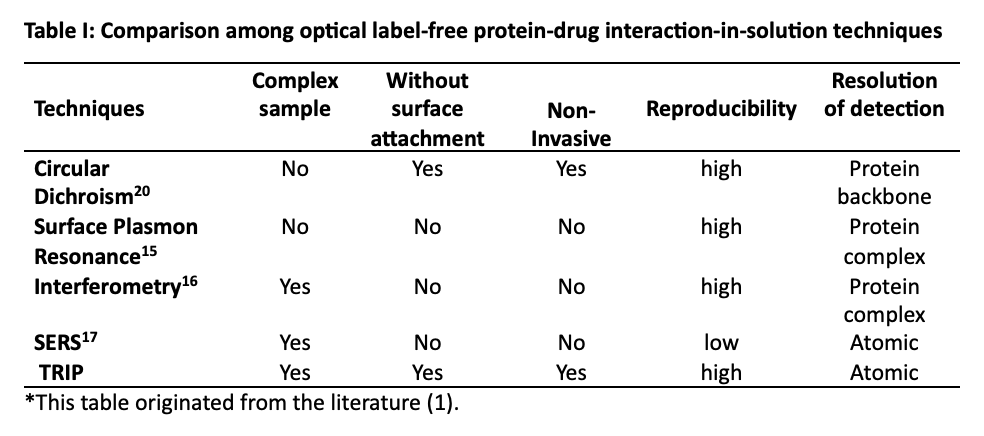
To see how the present TRIP technique compares with other optical approaches, we need only refer to Table I (1), which is, in part, given below:
What was unique about your study compared to previous studies done on this topic?
The TRIP technique offers several advantages that make it a cost-effective and efficient approach for high-throughput drug screening.
In Table I, we compare our technique to other label-free molecular diagnostic tools. The list in Table I is not intended to be exhaustive or complete, but only sufficient to give an idea of how Raman fits into the existing suite of tools available to biologists and chemists. Examination of the table illustrates that TRIP excels in operation at near-physiological conditions as mentioned above. The comparison with other techniques establishes TRIP as a general advance-screening tool for a wide variety of protein–drug interactions.
We summarize some of the key points of our research on TRIP below.
a. Sample Preparation: The elimination of sample preparation steps reduces the time and resources required for experiments. Researchers can directly analyze samples without the need for complex and time-consuming preparation protocols.
b. Minimal Protein Usage: TRIP uses diluted protein samples, which can be especially important when working with expensive or limited protein resources. This not only reduces costs, but it also conserves valuable research materials.
c. Rapid Detection: The ability to achieve rapid detection (within minutes) is a significant time-saver. Traditional drug screening methods can be time-intensive, whereas TRIP provides rapid results, allowing for more experiments to be conducted in a shorter period.
d. Direct Binding Measurement: TRIP directly measures binding interactions between proteins and ligands. This direct measurement avoids the need for secondary assays or readouts, simplifying the experimental process and reducing costs associated with additional assays.
e. Detection of time-dependent binding: Capability of time-dependent measurement of TRIP successfully demonstrated for the binding interactions between the transthyretin (TTR) and 2,4 dinitrophenol DNP molecules in Figure 2 (which can be seen in the literature [1]).
f. Low-Cost Extension: If TRIP can be implemented as an extension to existing systems, it minimizes the need for significant capital investments in new equipment. This cost-effective integration into existing laboratory setups makes it accessible to a broader range of research facilities.
These advantages collectively make TRIP an attractive option for high-throughput drug screening. It not only accelerates the screening process, but it also reduces the overall costs associated with drug discovery efforts. This is particularly valuable in pharmaceutical research, where the efficient identification of potential drug candidates can have a profound impact on the development of new therapies.
Usually, traditional drug screening techniques are only effective if the experimental conditions don’t influence them. What steps did you and your team need to ensure that your screening technique would not be influenced by experimental conditions?
We carefully addressed experimental conditions such as temperature, pH, buffer composition, laser power, and acquisition time across all experiments. This helped ensure that results are due to the interactions being studied rather than experimental artifacts. Performing experiments to replicate and assess the reproducibility of results is confirmed in clustering of the same protein in the PCA plots.We conducted control experiments with known protein-ligand interactions such as binding between streptavidin biotin and the bindings between protein A and antibodies to establish baseline measurements and assess the technique's accuracy. Also, we included the interactions of the SARS CoV-2 receptive binding domain (RBD) with non-binding and binding antibodies for checking positive and negative interactions.
Your study also used principal component analysis (PCA), stating that the method helped you and your team understand factors that affect spectral variation in the data you collected (1). Can you elaborate on this part of the study and explain how PCA assisted you in understanding your data?
Indeed, PCA is a valuable tool for understanding complex spectral data collected through techniques like Raman microscopy in our study. It simplifies data interpretation, aids in identifying influential variables, reduces noise, and facilitates the visualization and exploration of spectral variations. For example, spectral data collected using techniques like Raman microscopy are high-dimensional, with many variables (wavenumbers). PCA allows us to reduce the dimensionality of the data by identifying the most important directions (principal components) in the data space. These principal components are linear combinations of the original variables and capture the most significant sources of variation in the data. It helped us uncover meaningful insights and patterns within our data, contributing to a better understanding of the factors influencing molecular interactions and the performance of the TRIP technique.
What are the next steps in your work? What are a few questions this study raises that should be explored in a future study?
Research efforts are currently underway to employ alternative multivariate statistical techniques for the analysis of TRIP data. These approaches aim to facilitate the estimation of amino acid compositions and the determination of secondary protein structures across a wide range of protein sizes. Furthermore, we are carrying out TRIP studies involving a viral protein in the presence of four different drugs.
Reference
(1) Altangerel, N.; Neuman, B. W.; Hemmer, P. R.; Yakovlev, V. V.; Rajil, N.; Yi, Z. Sokolov, A. V.; Scully, M. O. Label-free drug interaction screening via Raman microscopy. PNAS 2023, 120, e2218826120. DOI: 10.1073/pnas.2218826120

Nanometer-Scale Studies Using Tip Enhanced Raman Spectroscopy
February 8th 2013Volker Deckert, the winner of the 2013 Charles Mann Award, is advancing the use of tip enhanced Raman spectroscopy (TERS) to push the lateral resolution of vibrational spectroscopy well below the Abbe limit, to achieve single-molecule sensitivity. Because the tip can be moved with sub-nanometer precision, structural information with unmatched spatial resolution can be achieved without the need of specific labels.