Absolute Quantification of Proteins and Peptides by ICP-MS
Special Issues
Despite all of the recent advances in analytical technologies dedicated to biotherapeutics, accurate protein quantification remains a challenge for the biopharmaceutical industry. UV spectrophotometry is commonly used for batch testing, but it requires the knowledge of the extinction coefficient of the protein, whose experimental determination requires the accurate concentration of a reference standard obtained by an absolute quantification method. To address the need for a fast analytical method capable of accurately quantifying a protein without any specific reference substance, an isotope dilution ICP-MS method was developed and validated, based on sulfur determination, allowing very accurate determination of a single protein in solution after microwave digestion.
The determination of accurate protein concentration is a challenge for biopharmaceutical companies when no reference standard is available. The commonly used techniques such as amino acid analysis are time consuming and suffer from poor precision and accuracy. An isotope dilution inductively coupled plasma–tandem mass spectrometry (ICP-MS/MS) method based on sulfur determination was developed and validated. It allows very accurate determination of a single protein in solution after microwave digestion. The method was validated using a National Institute of Standards and Technology (NIST) certified bovine serum albumin (BSA) solution with precision <1% relative standard deviation (RSD) and accuracy <2% bias over the range of concentrations tested.
Despite all of the recent advances in analytical technologies dedicated to biotherapeutics, accurate protein quantification remains a challenge for the biopharmaceutical industry. Ultraviolet (UV) spectrophotometry is commonly used for batch testing, but it requires the previous determination of the extinction coefficient of the protein. The accuracy of this determination is limited by the knowledge of the accurate concentration of a standard solution, again requiring an absolute quantification method.
Most protein quantification techniques (separative techniques such as liquid chromatography [LC] or capillary electrophoresis [CE], colorimetric assays, immunoassays, and so forth) cannot be considered as absolute because they also require a reference standard.
Amino acid analysis after complete hydrolysis of a protein is probably the most commonly used method for the absolute quantification of a single protein or peptide. However, hydrolysis and derivatization are time-consuming procedures and very often result in low precision and accuracy.
To address the need for a fast analytical method capable of accurately quantifying a protein without any specific reference substance, an isotope dilution inductively coupled plasma–tandem mass spectrometry (ICP-MS/MS) method based on sulfur determination was developed and validated, allowing for very accurate determination of a single protein in solution after microwave digestion.
Principle of Protein Quantification by ICP-MS
ICP-MS is mainly used to analyze metals in a wide range of applications, from environmental analysis to quality control of pharmaceuticals and biopharmaceuticals. However, it can also be used in biotechnology and proteomics thanks to its ability to quantify elements such as phosphorus.
Only a minority of proteins contain a metal ion or phosphorus in their composition, which prevents the use of ICP-MS for their quantification. However, sulfur is present in most proteins because it is contained in amino acids such as cysteine and methionine (1).
Sulfur determination by ICP-MS has long been a challenge because of the high ionization potential of this element (10.4 eV) and the interferences from polyatomic ions for all sulfur isotopes. Triple-quadrupole ICP-MS can easily circumvent these issues, as exemplified in Figure 1 for the determination of 32S+. By knowing the theoretical amount of sulfur in a given protein (from its amino acid sequence), sulfur determination can therefore lead to the absolute quantification of the protein in the sample.
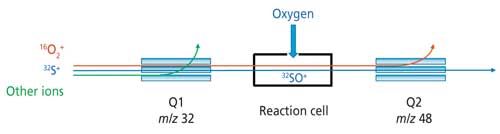
Figure 1: Principle of triple-quadrupole ICP-MS operating in mass-shift mode using oxygen as the reaction cell-gas for sulfur determination.
Most applications of ICP-MS for protein quantification involve the use of a chromatographic separation (2–4). This approach usually leads to under-estimation of the protein concentration if absorption occurs on the chromatographic system, which is often the case for hydrophobic proteins. In this study, microwave-assisted digestion was used for sample preparation and isotope dilution for quantification. This method was validated according to the International Conference on Harmonization (ICH) guideline Q2(R1) “Validation of Analytical Procedures: Text and Methodology” (6) using bovine serum albumin (BSA) standard reference material from the National Institute of Standards and Technology (NIST).
A similar approach was developed simultaneously by Lee and colleagues (5).
Materials and Methods
Equipment
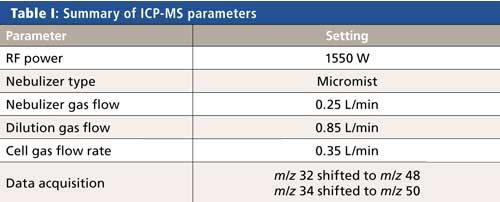
All analyses were carried out with an Agilent 8800 ICP-QQQ system fitted with a standard concentric nebulizer, x-lens, a Peltier-cooled double-pass Scott-type spray chamber, and standard Pt interface cones. Q1 was set to select ions 32S+ (m/z 32) and 34S+ (m/z 34), and Q2 was set to select 32S16O+ (m/z 48) and 34S16O+ (m/z 50) (mass shifted because of the reaction with oxygen in the reaction cell). A summary of ICP-MS parameters is presented in Table I. An Agilent I-AS integrated autosampler was used for sample infusion.
Digestion was performed in a Milestone UltraWAVE single reaction chamber microwave digestion system.
Sample Preparation
A protein sample containing approximately 50 µg of sulfur was weighed into a disposable glass tube. Then 50 µg of 34S was added (as H234SO4), followed by 2 mL of 69% HNO3, 0.5 mL of 37% HCl, and 1 mL of 30% H2O2. After microwave digestion, the sample was diluted to 50 mL with water. The same procedure was performed on 50 µg of NIST-traceable SO42- reference solution for reverse isotope dilution MS (vide supra).
To take into account the presence of sulfur-containing species in the formulation buffer of the protein, it is necessary to correct for the nonproteinaceous sulfur. To do so, the sample is filtered through a 3-kDa molecular-weight cut-off membrane and an aliquot of the filtrate is digested as a sample and serves as a blank.
Double Isotope Dilution Methodology
In a classical isotope dilution MS strategy, a known amount of an isotope-enriched standard solution is added to the sample (containing mainly the major isotope). The quantification is then performed by measuring the ratio between the two isotopes. In the method we developed, the spiking solution contains 34SO42- that is formed by oxidation of 34S powder. Thus, the accurate concentration of 34SO42- added to the sample is not known “as is.” Therefore, a double isotope dilution MS strategy was used: The concentration of the 34SO42- solution was determined by reversed isotope dilution MS with a NIST-traceable certified solution of sulfate. This approach also allows us to compensate for sensitivity differences observed between 32S and 34S (mass discrimination).
The equation used for the calculation of sulfur mass fraction wx (in micrograms per gram) in the sample is as follows:
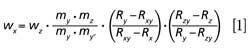
where x refers to the sample; y and y′ refer to 34SO42- spiking solution; z refers to the NIST SO42- reference solution; wz is the sulfur mass fraction in the NIST SO42- reference solution; mi is the mass of the sample, standard, or spiking solution; Ri is the isotopic ratio 34S–32S in the sample, standard, or spiking solutions; Rxy is the ratio measured when mx is spiked with my′; and Rzy is the ratio measured when mz is spiked with my′.
Method Validation
The method was validated according to the ICH Q2(R1) guideline (6), as regards to linearity of the method, accuracy, range, precision (repeatability and intermediate precision), robustness, and solutions stability. Specificity was assessed based on results from accuracy and robustness.
BSA was used as a model compound. To be able to assess the method accuracy, the validation was performed on NIST standard reference material (SRM) 927e-bovine serum albumin (67.38 ± 1.38 mg/mL). Method precision was also assessed for a monoclonal antibody sample (trastuzumab, an anti-HER2 antibody approved for the treatment of breast cancers).
Linearity of the Method and Accuracy
The linearity of the method and its accuracy were evaluated by digesting different amounts of BSA standard solution (NIST SRM 927e) in the presence of a constant amount of added 34S. Protein amounts corresponding to ~30, 40, 50, 60, and 70 µg of sulfur per sample were used. At each level, the analysis was performed in triplicate (three independent preparations). The graph for linearity assessment is presented in Figure 2. Mean recoveries ranged between 101.2% and 101.3%. Relative standard deviations (RSDs) at each level ranged between 0.1% and 0.3%. A linear relationship was obtained between the amount of BSA in the digestion tube and the amount of BSA experimentally measured (R = 1.000). The method is linear and accurate in the range 60–140% (100% level corresponds to 50 µg of sulfur in the digestion tube).
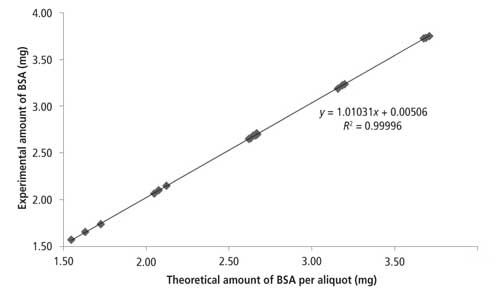
Figure 2: Method linearity assessment.
Method Precision
Method precision was assessed for two samples: BSA standard solution (NIST SRM 927e) and trastuzumab monoclonal antibody solution. Two analysts performed six determinations of each sample on two days. RSDs for each analyst (n = 6) ranged between 0.1% and 0.8%. The overall RSDs (n = 12) were below 0.7%. The relative differences between the means obtained by the two analysts were ≤1.4%. These results are within the criteria commonly accepted in the pharmaceutical industry for the validation of quantitative methods. The method can therefore be considered precise.
Robustness
Robustness was assessed relative to two parameters: protein concentration or sample dilution and sample matrix.
Protein concentration in the sample can be an issue: The sample volume transferred into the digestion tube should ideally be 1.0 mL. If the sample is too concentrated, it may be diluted before digestion. On the other hand, for samples with protein concentrations corresponding to sulfur content below 30 µg/mL, volumes larger than 1.0 mL are required. This need for larger volumes can have an impact on the digestion reaction rates and can affect the accuracy of the protein determination.
Constant amounts of the BSA standard solution and isotope standard were diluted with different amounts of water before digestion and analyzed. Recoveries and RSDs were calculated. Mean recoveries ranged between 100.3% and 100.9%. RSDs at each level ranged between 0.1% and 0.3%. No trend in accuracy or precision was observed when increasing the volume. No carryover effects were observed either. Therefore, it can be concluded that the method is valid for sample solutions containing 6–70 µg/mL of sulfur, in volumes of 1–5 mL.
The digestion step and results obtained can be affected by the components of the sample matrix. To demonstrate the robustness of the method versus different matrices, a BSA standard and an isotope standard were digested in matrices covering a range of different formulation buffers commonly used for therapeutic proteins. The formulations tested and their compositions are listed in Table II.
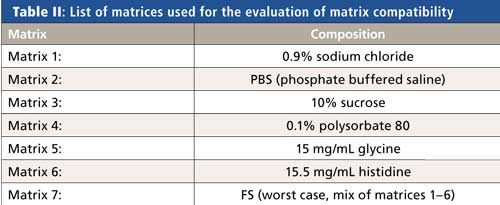
Protein concentration, precision of determination, and accuracy versus theoretical values were calculated. Mean recoveries ranged between 99.6% and 100.8% versus the theoretical value. RSDs (n = 3) ranged between 0.0% and 0.6%.
The performance of the method was not affected by the sample matrix for the formulations tested.
Determination of Nonproteinaceous Sulfur
To assess the accuracy of the nonproteinaceous sulfur determination, sulfur-containing low-molecular-weight compounds (methionine and sulfate ions) were added to BSA standard solution (NIST SRM 927e) and to a monoclonal antibody sample, at a concentration representing ~5% of the sulfur present in the protein. Samples were filtered through a 3-kDa molecular weight cut-off membrane and the sulfur concentration in the filtrate was measured by ICP-MS. The recoveries for nonproteinaceous sulfur ranged between 96.6% and 98.9%. This result shows that low-molecular-weight sulfur-containing compounds can be removed efficiently by filtration.
Stability of Solutions
Digested standard and sample solutions were stored in polypropylene tubes at room temperature for up to five days. Their stability was evaluated by comparison of sulfur concentrations (determined by isotope dilution) determined on different days (using different back-spike standardizations). Recoveries ranged between 99.6% and 100.8%. Solutions are usually considered stable if the difference observed on the result over the tested period is ≤2.0%-that is, a recovery between 98.0% and 102.0%. Stability of the solutions over five days was therefore demonstrated.
Conclusion
Double isotope dilution triple-quadrupole ICP-MS is an innovative method for the absolute quantification of pure proteins. The precision and accuracy of this methodology bear no comparison to the traditional methods used in most analytical laboratories, such as amino acid analysis. This methodology is tolerant to most matrices commonly used for therapeutic protein formulations, widely applicable, and fast (results can be obtained within half a day of work in routine use).
References
- M. Wind, A. Wegener, A. Eisenmenger, R. Kellner, and W.D. Lehmann, Angew. Chem. Int. Edit.42, 3425–3427 (2003).
- M. Wang, W. Feng, W. Lu, B. Li, B. Wang, M. Zhu, Y. Wang, H. Yuan, Y. Zhao, and Z. Chai, Anal. Chem. 79, 9128–9134 (2007).
- C. Rappel and D. Schaumlöffel, Anal. Bioanal. Chem. 390, 605–615 (2008).
- M. Wang, W. Feng, Y. Zhao, and Z. Chai, Mass Spectrom. Rev. 29, 326–348 (2010).
- H. Lee, S. Kim, J. Jeong, Y. Lee, and Y. Yim, Metrologia52, 619–627 (2015).
Juliusz Bianga, Géry Van Vyncht, Philippe De Raeve, and Arnaud Delobel are with Quality Assistance SA, in Donstiennes, Belgium. Direct correspondence to: arnaud.delobel@quality-assistance.be
