Calibrating the Composition of a Copolymer
In collaboration with Isao Noda, I have been using Raman spectroscopy to study the properties of the bioplastic polyhydroxybutyrate hexanoate (PHBHx), which depend on the percentage of hexanoate (which produces propyl side branches to the polymer chain). The percentages of hexanoate determine the maximum crystallinity that the polymer can experience, and that determines its physical and chemical properties, which include its optical clarity, dyability, flexibility, and thermal properties (melting and glass transition temperature). It is useful to have an easy way to determine the composition; in this column, we describe how Raman spectroscopy was used to show that this is feasible at an accuracy greater than expected.
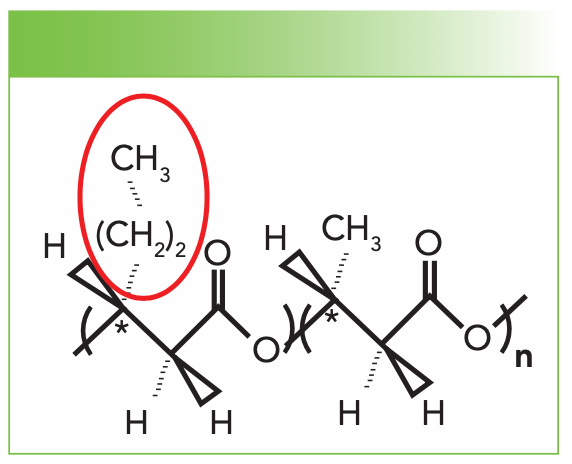
The structure of polyhydroxybutyrate hexanoate (PHBHx) is shown in Figure 1. Although the PHB homopolymer is highly crystalline, the insertion of the propyl group highlighted in red disrupts the ability of PHB to crystallize, resulting in a lower maximum crystallinity. The presence of the propyl group adds two methylene groups and can change the interaction of the methyl end group with the chain because it is separated from the chain by the two methylene groups. Thus, if one wants to quantitate the concentration of propyl units, it would be of interest to look in the region of the CH-stretching bands between 2700 and 3000 cm-1. However, this cannot be easily done in the solid phase because of the random, consistent errors that would be introduced by the spectral changes, which would stem from crystallinity and orientation. Thus, we can either measure the spectra in the melt or in the solution. Because the melting temperatures may vary, thus producing other ambiguities in the spectra, we opted to look at the solution phase. The obvious problem with the solution phase was the presence of the CH bands of the solvent. But, even then, the obvious solution to this problem would be to dissolve the samples in a deuterated solvent, which is what we did. PHBHx is soluble in chloroform (CHCl3), so we used deuterated chloroform (CDCl3).
FIGURE 1: The structure of PHBHx shows one propyl group incorporated into a chain of PHB in place of the methyl side group. The composition would be one propyl for every n methyl groups.
Description of Work Done
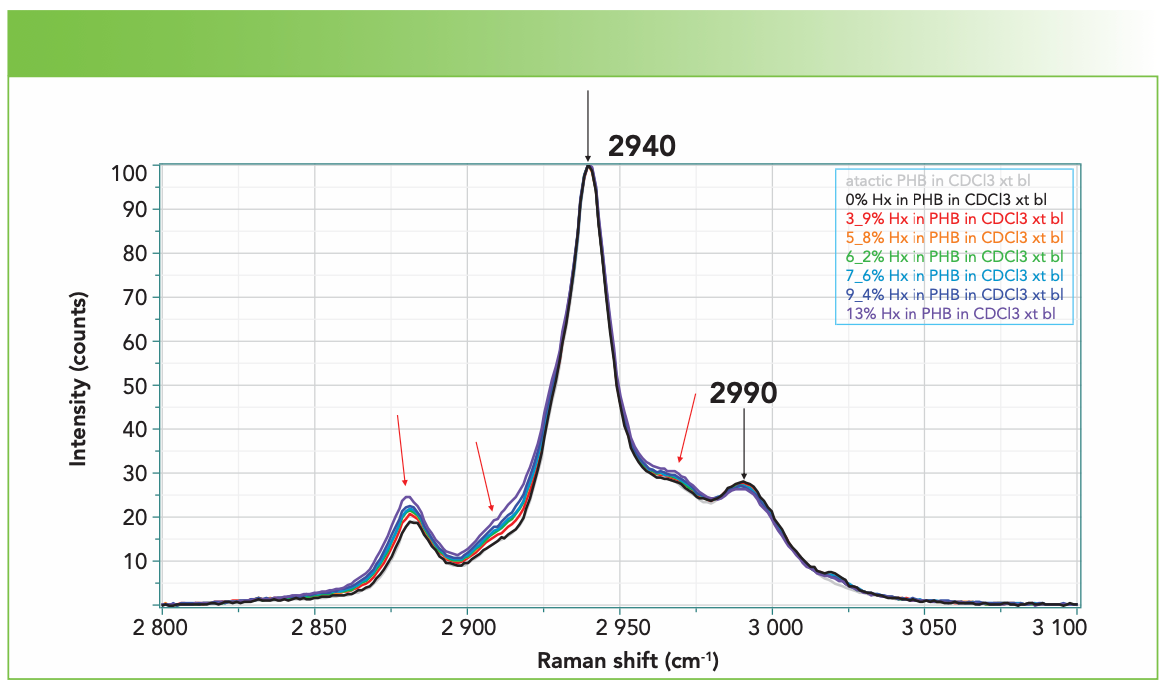
Figure 2 shows the spectra between 1900 and 3150 cm-1 collected on the XploRA using the 1800 groves/mm grating, a 532-nm laser, the 45° mirror enabling access to the sample vials with the 4-cm lens, and the wavelength fixed so that the CD stretch at 2254 cm-1 is observed simultaneously with the CH stretches of the polymer without scanning the monochromator, but still with good spectral resolution (~1.4 cm-1 per pixel, with the sharpest band being the CD stretch at less than 7 cm-1). Figure 2 is displayed with the CD stretch scaled the same in all spectra. Figure 3 shows the CH region extracted and scaled, so that the band at 2940 cm-1 is the same in all spectra. It was not clear a priori that the band at 2940 cm-1 would be the best band to use for normalization. Had the entire spectrum been acquired, we could have used the >C=O band whose concentration is independent of the substitution (refer to Figure 1). But, as you will see, the normalization we used produced good results.
FIGURE 2: The spectra of PHBHx with varying concentrations of Hx, from top to bottom, of atactic: 0%, 3.9%, 5.8%, 6.2%, 7.6%, 9.4%, and 13%. The spectra are displayed with the CD stretch at maximum intensity.
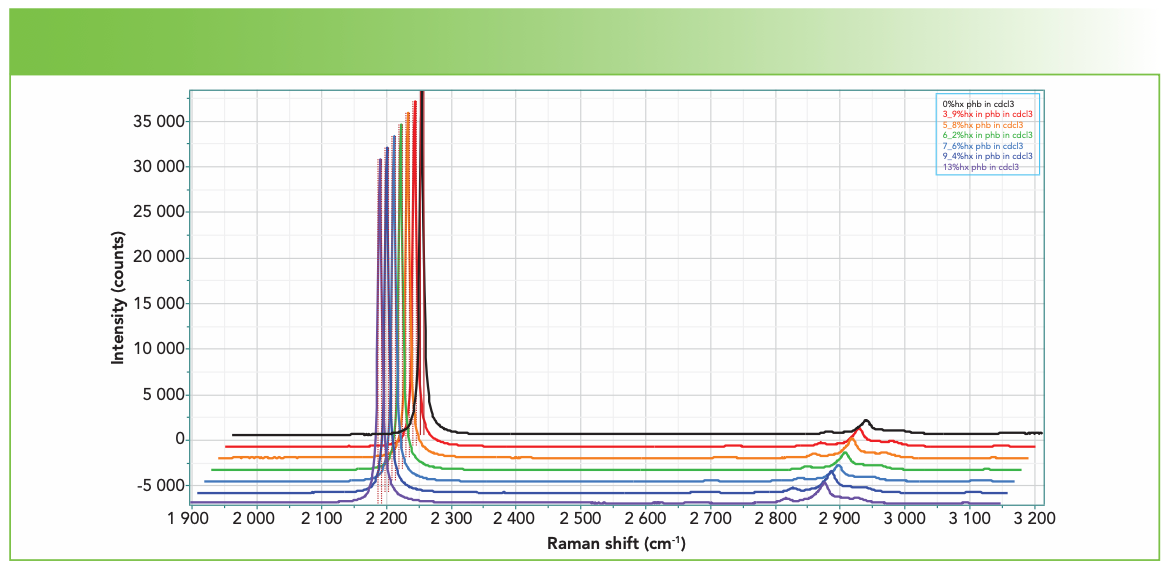
FIGURE 3: Spectra of the CH region of PHBHx with varying concentrations of Hx, from top to bottom, of atactic: 0%, 3.9%, 5.8%, 6.2%, 7.6%, 9.4%, and 13%, scaled to the maximum intensity band at 2940 cm-1. Small changes are indicated with arrows. The small feature at ~2920 cm-1 is an artifact.
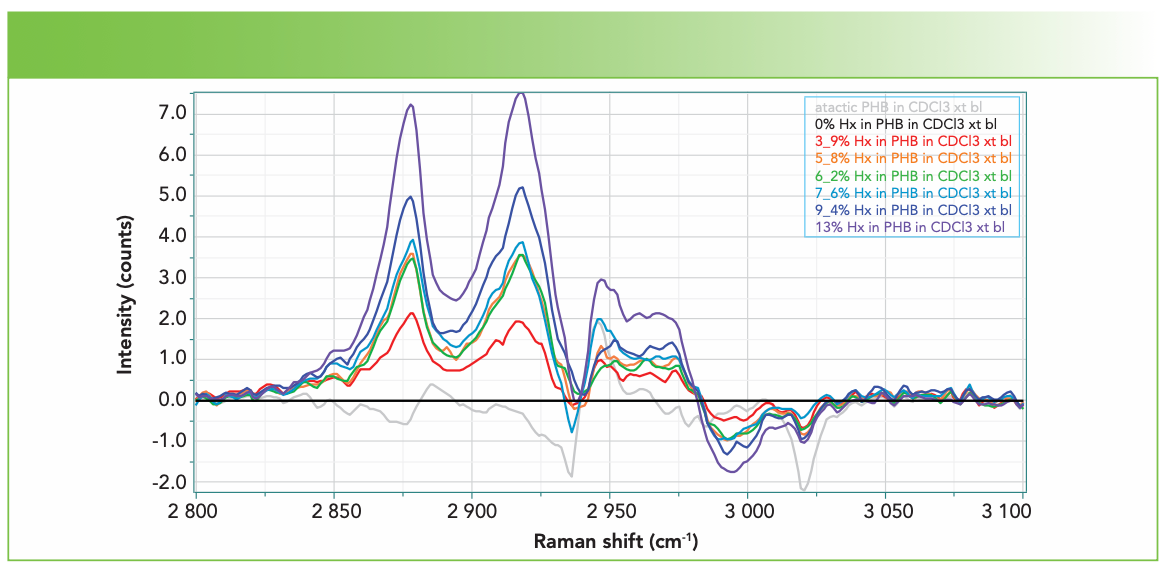
The next step that was performed before trying to produce a model for prediction was to subtract the spectrum of the 0% hexanoate sample (that is, PHB homopolymer). The result of this operation is shown on Figure 4. In support of this method, we see that the subtracted signals more or less scale with the percentage substitution (I color code the spectra in the sense of a rainbow to easily visualize the behavior).
FIGURE 4: Spectra of the CH region of PHBHx with varying concentrations of Hx, listed in the box on the upper right (from top to bottom), of atactic: 0%, 13%, 9.4%, 7.6%, 6.2%, 5.8%, 3.9%, and atactic, after subtraction of the 0% spectrum, which appears as the straight black line. These spectra have been smoothed after subtracting 0% Hx in PHB.
Using LabSpec to Produce a Model for Prediction
Now that I have a data set from a sample set with known comonomer percentages and I can see that the spectra are varying systematically with the percentages, it is time to try using partial least squares (PLS) to determine if a quantitative predictive model can be derived from the spectra.
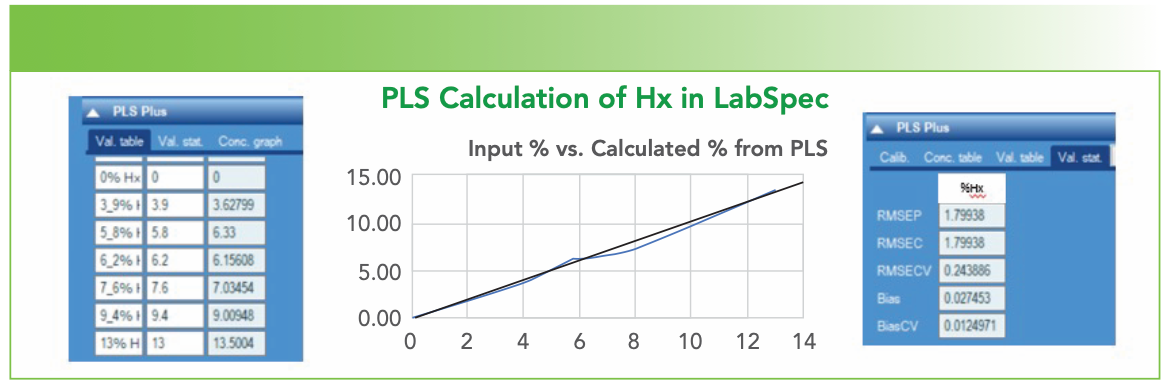
In LabSpec, this is easily done by importing a multifile with all of the relevant spectra. The user then inputs the values for the variable; in this case, the only variable was the percentage of Hx. The program automatically creates a model. Figure 5 shows the result of the model creation. The two blue tables were captured directly from LabSpec. The graph was created by simply copying the table on the left and pasting it into Excel. I have inserted the black line to indicate what a perfect fit would be (input value = calculated value). The plot in the middle was created in Excel because the x-axis in the LabSpec plot was in the spectrum number rather than the percentage. Curiously, Excel sometimes reported that the plot was correlated. I would hope so, since we would like the predicted value to be a good approximation of the input value!
FIGURE 5: Results of the PLS modeling in LabSpec. The plot in the middle was created in Excel.
Summary and Conclusion
Polymers have become extremely useful materials for a myriad of commercial applications. In particular, their properties can be engineered for the degree of crystallinity, which, in the case of PHBHx, depends on the concentration of the hexanoate. The sample set that was studied here has seven samples with Hx concentrations between 0 and 13%. In the manufacturing process, it is useful to confirm a material’s concentration before initiating the manufacturing of the final product with a large amount of material. The results shown here indicate that a simple Raman measurement can estimate the concentration of an unknown in the low percentage range to ±0.5%. The immediate implication for the product quality control (QC) during the commercial production of many thousands of pounds of the material is obvious.
Fran Adar is the Principal Raman Applications Scientist for HORIBA Scientific in Edison, New Jersey. Direct correspondence to: SpectroscopyEdit@mmhgroup.com. ●


Newsletter
Get essential updates on the latest spectroscopy technologies, regulatory standards, and best practices—subscribe today to Spectroscopy.
AI-Powered Raman with CARS Offers Laser Imaging for Rapid Cervical Cancer Diagnosis
July 15th 2025Chinese researchers have developed a cutting-edge cervical cancer diagnostic model that combines spontaneous Raman spectroscopy, CARS imaging, and artificial intelligence to achieve 100% accuracy in distinguishing healthy and cancerous tissue.
Nanometer-Scale Studies Using Tip Enhanced Raman Spectroscopy
February 8th 2013Volker Deckert, the winner of the 2013 Charles Mann Award, is advancing the use of tip enhanced Raman spectroscopy (TERS) to push the lateral resolution of vibrational spectroscopy well below the Abbe limit, to achieve single-molecule sensitivity. Because the tip can be moved with sub-nanometer precision, structural information with unmatched spatial resolution can be achieved without the need of specific labels.
sCMOS vs CCD: Advancing High-Speed CARS Spectroscopy
July 14th 2025A new comparative study shows that scientific CMOS (sCMOS) cameras could rival traditional CCD detectors in certain Raman CARS spectroscopy applications, offering faster readout and dynamic range despite slightly higher noise levels.