
Joanna Szpunar of the National Research Council of France discusses her use of ICP-MS methods to detect trace levels of human selenoproteins in cell extracts and for the identification of selenium containing proteins in rice.

Joanna Szpunar of the National Research Council of France discusses her use of ICP-MS methods to detect trace levels of human selenoproteins in cell extracts and for the identification of selenium containing proteins in rice.

Selenoproteins play an important role in human physiology and health, and as a result, sensitive methods are needed for their analysis. Joanna Szpunar of the National Research Council of France has been working with bioinorganic speciation analysis and hyphenated techniques for metallomics studies for some time. She recently spoke to Spectroscopy about using laser-ablation inductively coupled plasma–mass spectrometry (LA-ICP-MS) to detect trace levels of human selenoproteins in cell extracts and about her work with ICP-MS-assisted electrospray tandem MS for the identification of selenium-containing proteins in rice grown on seleniferous soils.

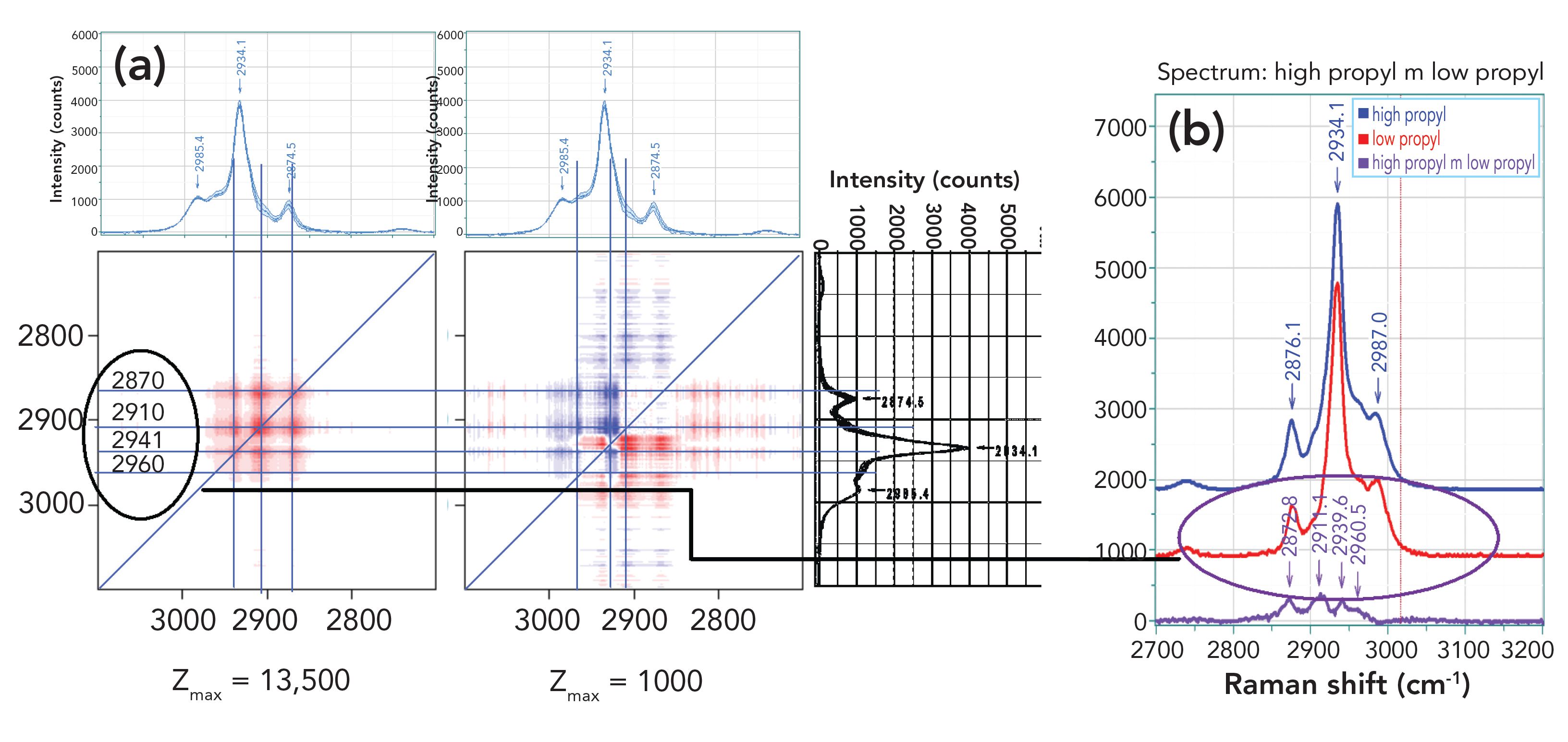
UV resonance Raman spectroscopy examines how UV light interacts with the electrons of samples and provides information about their molecular structure and dynamics. Sanford A. Asher, Distinguished Professor of Chemistry at the University of Pittsburgh, is using the technique to study peptide excited states and conformations and protein folding, with the ultimate goal of helping to advance research into the mechanisms of disease. He recently spoke to us about this work.


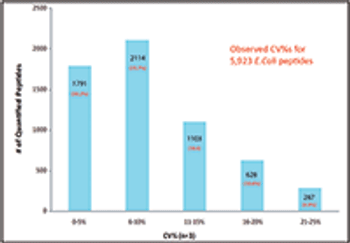
This article describes the development of a new data-independent acquisition (DIA) workflow for protein quantification that uses a mass spectrometer that combines three types of mass analyzers to achieve lower limits of detection (LOD), higher sensitivity, more accurate quantitative results, wider dynamic range, and better reproducibility than existing high-resolution accurate-mass (HRAM) tandem mass spectrometry (MS-MS) DIA workflows.
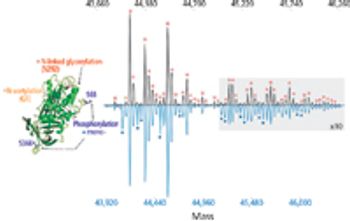
Native mass spectrometry, the method by which noncovalent protein complexes are retained in the gas phase for intact mass analysis, is gaining interest as a method for intact protein characterization. The development of a modified orbital ion trap platform for high-resolution analyses has expanded the role of native mass spectrometry to address the challenges of intact protein characterization.
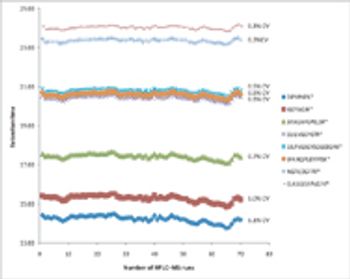
The detection limit, analytical precision, dynamic range, and robustness of a method for the targeted quantification of peptides using a capillary-flow LC–MS system were evaluated by spiking known amounts of isotopically labeled yeast peptides into a 500-ng yeast digest matrix.
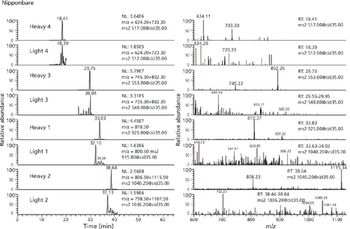
Liquid chromatography–mass spectrometry (LC–MS) successfully differentiated transgenic from native protein in a case where the proteins were highly homologous and could not be differentiated by traditional methods. This methodology may be useful for other studies of transgenic crops.
The inclusion of time-resolved selected reaction monitoring (SRM) functionalities into mass spectrometer control software allows large numbers of peptides to be quantified using short LC–MS-MS methods.
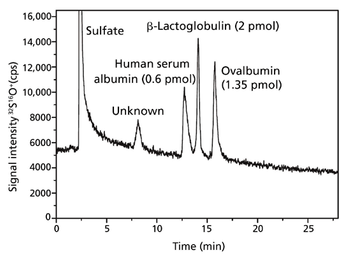
With recent research, the University of Oviedo's analytical spectrometry research group has taken a step closer to the absolute quantification of proteins. Quantification based upon isotope dilution mass spectrometry of sulfur is hampered by gas-based polyatomic interferences. By implementing a quadrupole inductively coupled mass spectrometer with collision/reaction cell technology, the group has been able to overcome the issues and has increased reliability while optimizing the efficiency of its analyses.
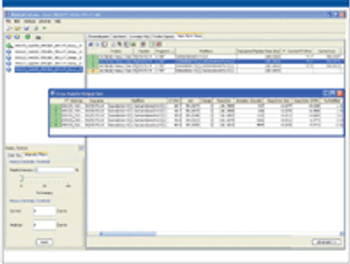
This article presents an efficient analytical workflow for protein characterization using LC–MS.
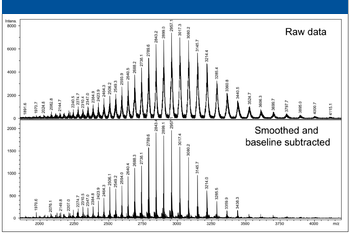
The author discusses the use of high-performance mass spectrometry for small molecule and protein applications.
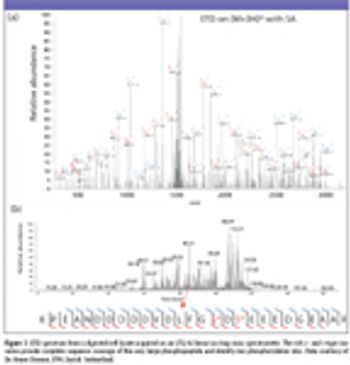
The authors discuss the use of electron transfer dissociation as a reliable tool in proteomic research, especially in conjunction with a linear ion trap, for sequence analysis of post-translationally modified and highly basic peptides.
The nanoLC LIT-TOF approach combines multiple capabilities that improve the ability to characterize complex protein mixtures significantly.
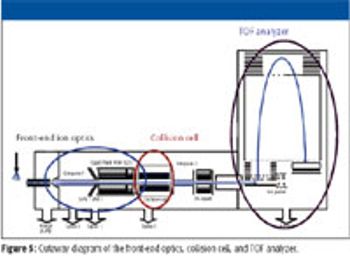
Because it is extremely rapid, biomarker discovery and identification using liquid chromatography–mass spectrometry (LC-MS), including both ion-trap and triple-quadrupole LC–MS, is well established. Fractionation of complex samples before LC–MS-MS analysis might be necessary to identify the proteins, greatly increasing the number of analyses required. In this case, there is ongoing debate regarding knowing whether the protein is identified correctly, knowing how much prior fractionation is needed to reduce complexity to the point where low-abundance proteins can be detected reliably, and balancing specificity with sensitivity.
